HDU1560 DNA sequence(IDA星) (E)
2017-07-09 10:02
507 查看
[align=left]Problem Description[/align]
The twenty-first century is a biology-technology developing century. We know that a gene is made of DNA. The nucleotide bases from which DNA is built are A(adenine), C(cytosine), G(guanine), and
T(thymine). Finding the longest common subsequence between DNA/Protein sequences is one of the basic problems in modern computational molecular biology. But this problem is a little different. Given several DNA sequences, you are asked to make a shortest sequence
from them so that each of the given sequence is the subsequence of it.
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.
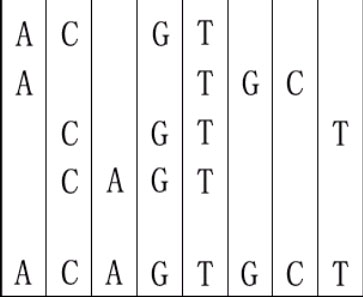
[align=left]Input[/align]
The first line is the test case number t. Then t test cases follow. In each case, the first line is an integer n ( 1<=n<=8 ) represents number of the DNA sequences. The following k lines contain
the k sequences, one per line. Assuming that the length of any sequence is between 1 and 5.
[align=left]Output[/align]
For each test case, print a line containing the length of the shortest sequence that can be made from these sequences.
[align=left]Sample Input[/align]
1
4
ACGT
ATGC
CGTT
CAGT
[align=left]Sample Output[/align]
8
题意就是给出N个DNA序列,要求出一个包含这n个序列的最短序列是多长
思路:就是IDA*迭代加深进行深搜,进行估价判断 f=g+h,g就相当于层数,h就是每个字符串还剩下的未匹配的长度的
每一层都对4个字符ACGT进行检验,如果能匹配到至少一个则继续往下深搜
#include<stdio.h>
#include<string.h>
#include<cstring>
#include<math.h>
#include<queue>
#include<stack>
#include<algorithm>
using namespace std;
int n;
struct node
{
char s[20];
int len;
}a[10];
int deep;
char s[10]="ACGT";
int pos[10];//记录每个字符串已经匹配到了哪个位置
int get_h()
{
int ans=0;
for(int i=0;i<n;i++)
{
ans=max(ans,a[i].len-pos[i]);//即剩余的还未匹配的字符串的长度
}
return ans;
}
bool dfs(int step)//step为从初始到当前状态的实际代价
{
if(step+get_h()>deep)return 0;//当前长度+估测的长度比deep还大的话,也就没有继续往下搜索的必要了
//f=g+h 估价函数
if(get_h()==0)return 1;
for(int i=0;i<4;i++)
{
int flag=0;//用于判断如果该位置放c[i]字符是否能匹配到相应的字符
int tem[10];
for(int j=0;j<n;j++)//记录 用于深搜之后的恢复
tem[j]=pos[j];
for(int j=0;j<n;j++)
{
if(a[j].s[pos[j]]==s[i])//当前这位符合,则该串的位置往后移一位
{
pos[j]++;
flag=1;
}
}
if(flag)//有符合的,则往下搜索
{
if(dfs(step+1))return 1;
for(int j=0;j<n;j++)//恢复
pos[j]=tem[j];
}
}
return 0;
}
int main()
{
int t;
scanf("%d",&t);
while(t--)
{
scanf("%d",&n);
int maxn=0;
for(int i=0;i<n;i++)
{
scanf("%s",a[i].s);
a[i].len=strlen(a[i].s);
maxn=max(maxn,a[i].len);
pos[i]=0;
}
deep
c29a
=maxn;
while(1)
{
if(dfs(0))break;
deep++;
}
printf("%d\n",deep);
}
return 0;
}
The twenty-first century is a biology-technology developing century. We know that a gene is made of DNA. The nucleotide bases from which DNA is built are A(adenine), C(cytosine), G(guanine), and
T(thymine). Finding the longest common subsequence between DNA/Protein sequences is one of the basic problems in modern computational molecular biology. But this problem is a little different. Given several DNA sequences, you are asked to make a shortest sequence
from them so that each of the given sequence is the subsequence of it.
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.

[align=left]Input[/align]
The first line is the test case number t. Then t test cases follow. In each case, the first line is an integer n ( 1<=n<=8 ) represents number of the DNA sequences. The following k lines contain
the k sequences, one per line. Assuming that the length of any sequence is between 1 and 5.
[align=left]Output[/align]
For each test case, print a line containing the length of the shortest sequence that can be made from these sequences.
[align=left]Sample Input[/align]
1
4
ACGT
ATGC
CGTT
CAGT
[align=left]Sample Output[/align]
8
题意就是给出N个DNA序列,要求出一个包含这n个序列的最短序列是多长
思路:就是IDA*迭代加深进行深搜,进行估价判断 f=g+h,g就相当于层数,h就是每个字符串还剩下的未匹配的长度的
每一层都对4个字符ACGT进行检验,如果能匹配到至少一个则继续往下深搜
#include<stdio.h>
#include<string.h>
#include<cstring>
#include<math.h>
#include<queue>
#include<stack>
#include<algorithm>
using namespace std;
int n;
struct node
{
char s[20];
int len;
}a[10];
int deep;
char s[10]="ACGT";
int pos[10];//记录每个字符串已经匹配到了哪个位置
int get_h()
{
int ans=0;
for(int i=0;i<n;i++)
{
ans=max(ans,a[i].len-pos[i]);//即剩余的还未匹配的字符串的长度
}
return ans;
}
bool dfs(int step)//step为从初始到当前状态的实际代价
{
if(step+get_h()>deep)return 0;//当前长度+估测的长度比deep还大的话,也就没有继续往下搜索的必要了
//f=g+h 估价函数
if(get_h()==0)return 1;
for(int i=0;i<4;i++)
{
int flag=0;//用于判断如果该位置放c[i]字符是否能匹配到相应的字符
int tem[10];
for(int j=0;j<n;j++)//记录 用于深搜之后的恢复
tem[j]=pos[j];
for(int j=0;j<n;j++)
{
if(a[j].s[pos[j]]==s[i])//当前这位符合,则该串的位置往后移一位
{
pos[j]++;
flag=1;
}
}
if(flag)//有符合的,则往下搜索
{
if(dfs(step+1))return 1;
for(int j=0;j<n;j++)//恢复
pos[j]=tem[j];
}
}
return 0;
}
int main()
{
int t;
scanf("%d",&t);
while(t--)
{
scanf("%d",&n);
int maxn=0;
for(int i=0;i<n;i++)
{
scanf("%s",a[i].s);
a[i].len=strlen(a[i].s);
maxn=max(maxn,a[i].len);
pos[i]=0;
}
deep
c29a
=maxn;
while(1)
{
if(dfs(0))break;
deep++;
}
printf("%d\n",deep);
}
return 0;
}
相关文章推荐
- hdu 1560 DNA sequence(IDA*)
- hdu 1560 DNA sequence(IDA*)
- HDU 1560 DNA sequence(IDA*搜索)
- HDU 1560 DNA sequence IDA*搜索
- HDU 1560 DNA sequence(IDA*)
- HDU 1560 - DNA sequence ( IDA* )
- Hdu1560 DNA sequence(IDA*)
- HDU 1560 DNA sequence(IDA*)
- HDU 1560 DNA sequence(IDA*~)
- HDU 1560 DNA sequence IDA*
- HDU-1560 DNA sequence(IDA*)
- hdu 1560 DNA sequence(迭代加深搜索)
- HDU - 1560----DNA sequence
- hdu 1560 DNA sequence(迭代加深搜索)
- HDU 1506 DNA sequence IDA*
- hdu 1560 DNA sequence(搜索)
- hdu1560 DNA sequence (迭代加深搜索)
- HDU-1560 DNA sequence
- HDU-#1560 DNA sequence(迭代加深搜)
- hdu1560 DNA序列(IDA*)
